From Arizona State University: DNA Provides Solution to our Enormous Data Storage Problem
ASU researchers show how molecular structures can store large volumes of data while providing powerful encryption
This is a Press Release edited by StorageNewsletter.com on February 9, 2026 at 2:00 pmBy Richard Harth, Biodesign Institute, Arizona State University
Since the dawn of the computer age, researchers have wrestled with 2 persistent challenges: how to store ever-increasing reams of data and how to protect that information from unintended access.
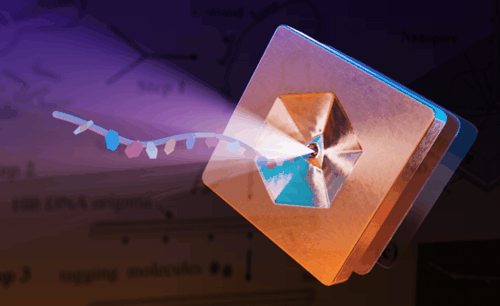
An illustration shows a strand of engineered DNA passing through a nanoscale sensor,
where its physical structure can be decoded as digital information.
DNA nanostructures could one day serve as ultra-dense carriers
of digital information and advance the field of data encryption.
(Graphic by Jason Drees/ASU)
Now, researchers with Arizona State University (ASU)’s Biodesign Institute and their colleagues offer a surprising answer. In a pair of new studies, they show how DNA, the molecule of life, can be harnessed to faithfully store enormous volumes of data and provide powerful encryption.
The findings, appearing in the journals Advanced Functional Materials and Nature Communications respectively, provide a nature-inspired alternative to silicon-based storage and encryption solutions. They could help reshape the design of future microelectronic and molecular information systems for a broad range of applications.
“For decades, information technology has relied almost entirely on silicon,” said Hao Yan, regents professor, School of Molecular Sciences and director, Biodesign Center for Molecular Design and Biomimetics, Arizona State University. “What we’re showing here is that biological molecules, specifically DNA, can be engineered to store and protect information in fundamentally new ways. By treating DNA as an information platform rather than just a genetic material, we can begin to rethink how data is stored, read and secured at the nanoscale.”
Yan, along with researchers Chao Wang, associate professor, School of Electrical, Computer and Energy Engineering, and Rizal Hariadi, associate professor, Department of Physics, worked together to lead the projects The team co-authored the 2 studies with other ASU collaborators. The 2 first authors of those papers are Gde Bimananda Mahardika Wisna and Penkun Xia..
Big data, tiny molecule
As the world generates tremendous volumes of digital information, today’s storage technologies are struggling to keep up. The first study demonstrates a new way to store information using DNA — not by analyzing genetic letters, but by interpreting DNA’s physical shape.
DNA is appealing because it can store massive amounts of information in a tiny physical volume, and because it can remain stable for astonishingly long periods. (In 2022, researchers recovered DNA fragments from Greenland sediments dating back roughly 2 million years.)
The new research describes the design and construction of tiny DNA structures that act like physical letters in an alphabet, each carrying a piece of information. As the structures pass through a microscopic sensor, machine learning software records and analyzes subtle electrical signals. Then, the system can translate the data back into readable words and short messages with high accuracy.
The approach offers a powerful alternative to more traditional DNA data storage methods that rely on slow and expensive DNA sequencing. In contrast, the new technique is faster, cheaper and more scalable.
The work points toward a future where DNA could serve as an ultra-dense, long-lasting and secure medium for data storage. It could be useful for archiving massive amounts of information — from scientific records to cultural data — using very little space and energy. It also demonstrates a powerful bridge between synthetic biology and semiconductor technology, opening the door to new kinds of molecular information systems that go beyond conventional electronics.
“By treating DNA as an information platform rather than just a genetic material, we can begin to rethink how data is stored, read and secured at the nanoscale“. said Hao Yan, Regents Professor, ASU.
Locking down information at the molecular level
While the 1st study focuses on how DNA can store information efficiently, the second explores how DNA nanostructures could also help protect information through encryption.
In this work, the researchers design intricate DNA origami structures — folded arrangements of DNA strands that form precise two- and three-dimensional shapes. Instead of storing data simply as bits or letters, information is encoded in the arrangement and pattern of these nanoscale structures. This creates a kind of molecular code that is difficult to interpret without the correct tools and reference patterns.
To read the encrypted information, the team uses an advanced form of super-resolution microscopy that can visualize individual DNA structures at extremely high precision. Machine learning software then analyzes thousands of molecular images, grouping similar patterns and translating them back into the original message. Without the correct decoding framework, the patterns are essentially meaningless, adding a layer of built-in security.
The approach greatly increases the number of possible molecular codes that can be created, making unauthorized decoding far more difficult. It also allows information to be packed into three-dimensional DNA structures, which adds even more complexity and security to each molecular key.
“In these studies, our team brings together complementary approaches, including DNA nanotechnology, super-resolution optical imaging, high-speed electronic readout and machine learning, to interrogate DNA nanostructures across multiple spatial and temporal scales,” Wang said. “This integrated strategy helps us better understand the behavior and function of DNA nanostructures.
“This is a very good example of doing research at the intersection of semiconductor technology and biology. In this emerging field, more applications, from advanced biosensing to programmable nanodevices, remain to be explored.”
Bringing storage and security together at the molecular scale
Together, the 2 studies studies (both studies were funded by the National Science Foundation’s Semiconductor Synthetic Biology Circuits and Communications for Information Storage (SemiSynBio) program), show how DNA can function not only as a compact storage medium, but also as a platform for secure information handling at the nanoscale. One technique emphasizes fast, electronic-style readout of molecular information, while the other demonstrates how molecular patterns themselves can serve as encrypted carriers of data.
DNA-based systems could one day support ultra-dense storage for scientific data, medical records or cultural archives. Molecular encryption could provide new ways to secure sensitive information in environments where conventional electronics struggle, such as extreme temperatures, radiation or long-term preservation.
The research highlights a growing convergence between biology, materials science, computation and electronics. By treating DNA as both a biological molecule and an information platform, researchers are opening new ways to store, protect and access data at scales far smaller and potentially far more durable than today’s digital devices.
Resource:
Related research: ASU researchers discover DNA-based electronic storage system
Article: High-speed 3D DNA PAINT and unsupervised clustering for unlocking 3D DNA origami cryptography
Findings show that DNA-based cryptography is a secure and versatile solution for storing information.
Nature Communications has published an article written by Gde Bimananda Mahardika Wisna, Department of Physics, Arizona State University, Tempe, AZ, USA, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and Center for Biological Physics, Arizona State University, Tempe, AZ, USA, Daria Sukhareva, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and School of Molecular Sciences, Arizona State University, Tempe, AZ, USA, Jonathan Zhao, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and School of Computing and Augmented Intelligence, Arizona State University, Tempe, AZ, USA, Prathamesh Chopade, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, Deeksha Satyabola, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and School of Molecular Sciences, Arizona State University, Tempe, AZ, USA, Michael Matthies, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, Subhajit Roy, Department of Physics, Arizona State University, Tempe, AZ, USA, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and Center for Biological Physics, Arizona State University, Tempe, AZ, USA, Chao Wang, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, USA, Petr Šulc, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, Center for Biological Physics, Arizona State University, Tempe, AZ, USA, and School of Molecular Sciences, Arizona State University, Tempe, AZ, USA, Hao Yan, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and School of Molecular Sciences, Arizona State University, Tempe, AZ, USA, and Rizal F. Hariadi, Department of Physics, Arizona State University, Tempe, AZ, USA, Center for Molecular Design and Biomimetics at the Biodesign Institute, Arizona State University, Tempe, AZ, USA, and Center for Biological Physics, Arizona State University, Tempe, AZ, USA.
Abstract: “DNA origami information storage is a promising alternative to silicon-based data storage, offering a molecular cryptography technique concealing information within DNA origami. Routing, sliding, and interlacing staple strands lead to a large 700-bit key size. Practical DNA data storage requires high information density, robust security, and accurate and rapid information retrieval. Consequently, advanced readout techniques and large encryption key sizes are essential. Here, we report an enhanced DNA origami cryptography protocol in 2D and 3D DNA origami, increasing the encryption key size. We employ all-DNA-based steganography with fast readout through high-speed DNA-PAINT super-resolution imaging. By combining DNA-PAINT data with unsupervised clustering, we achieve an accuracy of up to 89%, despite the flexibility in the 3D DNA origami shown by oxDNA simulation. Furthermore, we propose criteria that ensure complete information retrieval for the DNA origami cryptography. Our findings show that DNA-based cryptography is a secure and versatile solution for storing information.“
Article: DNA Helix Bundle-Encoded Multi-Bit Information Readout by Sapphire-Supported Nanopores
Work highlights the potential of integrating DNA nanostructures with low-noise nanopore technology for high-density, secure, and scalable data storage applications.
Advanced Functional Materials has published an article written by Pengkun Xia, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, Deeksha Satyabola, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and School of Molecular Sciences, Arizona State University, Tempe, Arizona, 85287 USA, Nimarpreet Kaur Bamrah, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, Md Ashiqur Rahman Laskar, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, Abdulla Al Mamun, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, Xu Zhou, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, Jiangsu, 210023 China, Gde Bimananda Mahardika Wisna, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and Center for Biological Physics and Department of Physics, Arizona State University, Tempe, AZ, 85287 USA, Yinan Zhang, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and School of Chemical Science and Engineering, Tongji University, Shanghai, 200092 China, Ashif Ikbal, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, Andrew Kemeklis, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, and Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, Alexandra E Krylova, Department of Biomedical Engineering, the University of Texas at Austin, Austin, TX, 78712 USA, Rizal F. Hariadi, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and Center for Biological Physics and Department of Physics, Arizona State University, Tempe, AZ, 85287 USA, Hao Yan, Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA, and School of Molecular Sciences, Arizona State University, Tempe, Arizona, 85287 USA, and Chao Wang, School of Electrical, Computer and Energy Engineering, Arizona State University, Tempe, AZ, 85287 USA, Center for Photonics Innovation, Arizona State University, Tempe, AZ, 85287 USA, and Biodesign Center for Molecular Design & Biomimetics, Arizona State University, Tempe, AZ, 85287 USA.
Abstract: “The increasing demand for high-capacity data storage continues to drive innovation in memory technologies. DNA has recently emerged as a promising medium due to its exceptionally high data density and long-term stability. Here, a DNA-based data storage and readout scheme is presented that leverages sapphire-supported, low-noise solid-state nanopores for reliable detection of nanostructured DNA. Data are encoded onto four-helix bundle (4HB) DNA structures, enabling flexible programmability and secure readout. The sapphire-supported nanopores demonstrate a high signal-to-noise ratio (≈22) under a 250 kHz filtering frequency, allowing precise signal discrimination during readout. Using a decision-tree supervised learning algorithm, up to ≈93% of encoded messages are accurately classified, demonstrating the feasibility of letter and message encoding for information retrieval. This work highlights the potential of integrating DNA nanostructures with low-noise nanopore technology for high-density, secure, and scalable data storage applications.“









 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

