For Eindhoven University of Technology, Future of Storage Lies in DNA Microcapsules
DNA archival storage within reach thanks to new Polymerase Chain Reaction technique
This is a Press Release edited by StorageNewsletter.com on May 10, 2023 at 2:01 pmStoring data in DNA sounds like science fiction, yet it lies in the near future.
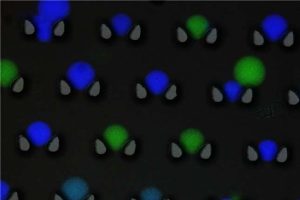
Microcapsules with fluorescent labels
Photo: Tom de Greef
Professor Tom de Greef expects the first DNA data center to be up and running within 5 to 10 years. Data won’t be stored as zeros and ones in a hard drive but in the base pairs that make up DNA: AT and CG. Such a data center would take the form of a lab, many times smaller than the ones today. De Greef can already picture it all. In one part of the building, new files will be encoded via DNA synthesis. Another part will contain large fields of capsules, each capsule packed with a file. A robotic arm will remove a capsule, read its contents and place it back.
We’re talking about synthetic DNA. In the lab, bases are stuck together in a certain order to form synthetically produced strands of DNA. Files and photos that are currently stored in data centers can then be stored in DNA. For now, the technique is suitable only for archival storage. This is because the reading of stored data is very expensive, so you want to consult the DNA files as little as possible.
Tom de Greef
Photo: Bart van Overbeeke
Large, energy-guzzling data centers made obsolete
Data storage in DNA offers many advantages. A DNA file can be stored much more compactly, for instance, and the lifespan of the data is also many times longer. But perhaps most importantly, this new technology renders large, energy-guzzling data centers obsolete.
And this is desperately needed, warns De Greef, “because in three years, we will generate so much data worldwide that we won’t be able to store half of it.”
Together with PhD student Bas Bögels, Microsoft Corp. and a group of university partners, he has developed a new technique to make the innovation of data storage with synthetic DNA scalable. The results have been published in the journal Nature Nanotechnology. De Greef works at the Department of Biomedical Engineering and the Institute for Complex Molecular Systems (ICMS) at TU Eindhoven and serves as a visiting professor at Radboud University.
Scalable
The idea of using strands of DNA for storage emerged in the 1980s but was far too difficult and expensive at the time. It became technically possible 3 decades later, when DNA synthesis started to take off. George Church, geneticist, Harvard Medical School, elaborated on the idea in 2011. Since then, synthesis and the reading of data have become exponentially cheaper, finally bringing the technology to the market.
In recent years, De Greef and his group have looked mainly into reading the stored data. For the time being, this is the biggest problem facing this new technique. The PCR method currently used for this, called ‘random access’, is highly error-prone. You can therefore only read one file at a time and, in addition, the data quality deteriorates too much each time you read a file. Not exactly scalable.
Here’s how it works: PCR (Polymerase Chain Reaction) creates millions of copies of the piece of DNA that you need by adding a primer with the desired DNA code. Corona tests in the lab, for example, are based on this: even a minuscule amount of coronavirus material from your nose is detectable when copied so many times. But if you want to read multiple files simultaneously, you need multiple primer pairs doing their work at the same time. This creates many errors in the copying process.
Every capsule contains one file
This is where the capsules come into play. De Greef’s group developed a microcapsule of proteins and a polymer and then anchored one file per capsule.
De Greef: “These capsules have thermal properties that we can use to our advantage.”
Above 50°C, the capsules seal themselves, allowing the PCR process to take place separately in each capsule. Not much room for error then. De Greef calls this ‘thermo-confined PCR’. In the lab, it has so far managed to read 25 files simultaneously without significant error.
If you then lower the temperature again, the copies detach from the capsule and the anchored original remains, meaning that the quality of your original file does not deteriorate.
De Greef: “We currently stand at a loss of 0.3% after three reads, compared to 35% with the existing method.”
Searchable with fluorescence
And that’s not all. He has also made the data library even easier to search. Each file is given a fluorescent label and each capsule its own color. A device can then recognize the colors and separate them from one another. This brings us back to the imaginary robotic arm at the beginning of this story, which will neatly select the desired file from the pool of capsules in the future.
This solves the problem of reading the data. De Greef: “Now it’s just a matter of waiting until the costs of DNA synthesis fall further. The technique will then be ready for application.”
As a result, he hopes that the Netherlands will soon be able to open its inaugural DNA data center – a world first.
This paper appeared in the journal Nature Nanotechnology under the title ‘DNA storage in thermoresponsive microcapsules for repeated random multiplexed data access’. DOI: 10.1038/s41565-023-01377-4.
Industrial partners:Microsoft Corp.
University partners: University of Washington, Radboud University, University of Bristol, Shanghai Jiao Tong University.
Partnerships: Center for Living Technologies, Eindhoven-Wageningen-Utrecht Alliance.
Article: DNA storage in thermoresponsive microcapsules for repeated random multiplexed data access
Nature Nanotechnology has published an article written by Bas W. A. Bögels, Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands, Computational Biology Group, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Bichlien H. Nguyen, Microsoft, Redmond, WA, USA, and Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, USA, David Ward, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, USA , Levena Gascoigne, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands, and Laboratory of Self-Organizing Soft Matter, Department of Chemical Engineering and Chemistry, Eindhoven University of Technology, Eindhoven, The Netherlands, David P. Schrijver, Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Anna-Maria Makri Pistikou, Alex Joesaar, Shuo Yang, Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands, and Computational Biology Group, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Ilja K. Voets, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands, and Laboratory of Self-Organizing Soft Matter, Department of Chemical Engineering and Chemistry, Eindhoven University of Technology, Eindhoven, The Netherlands, Willem J. M. Mulder, Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, and Department of Internal Medicine and Radboud Center for Infectious Diseases (RCI), Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands, Andrew Phillips, Microsoft Research, Cambridge, UK, Stephen Mann, Centre for Protolife Research and Centre for Organized Matter Chemistry, School of Chemistry, University of Bristol, Bristol, UK, School of Materials Science and Engineering, Shanghai Jiao Tong University, Shanghai, People’s Republic of China, and Zhangjiang Institute for Advanced Study (ZIAS), Shanghai Jiao Tong University, Shanghai, People’s Republic of China, Georg Seelig, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, USA, and Department of Electrical Engineering, University of Washington, Seattle, WA, USA, Karin Strauss, Yuan-Jyue Chen, Microsoft, Redmond, WA, USA, and Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, WA, USA, Tom F. A. de Greef, Laboratory of Chemical Biology, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Institute for Complex Molecular Systems (ICMS), Eindhoven University of Technology, Eindhoven, The Netherlands, Computational Biology Group, Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, The Netherlands, Institute for Molecules and Materials, Radboud University, Nijmegen, The Netherlands, and Center for Living Technologies, Eindhoven-Wageningen-Utrecht Alliance, Utrecht, The Netherlands.
Abstract: “DNA has emerged as an attractive medium for archival data storage due to its durability and high information density. Scalable parallel random access to information is a desirable property of any storage system. For DNA-based storage systems, however, this still needs to be robustly established. Here we report on a thermoconfined polymerase chain reaction, which enables multiplexed, repeated random access to compartmentalized DNA files. The strategy is based on localizing biotin-functionalized oligonucleotides inside thermoresponsive, semipermeable microcapsules. At low temperatures, microcapsules are permeable to enzymes, primers and amplified products, whereas at high temperatures, membrane collapse prevents molecular crosstalk during amplification. Our data show that the platform outperforms non-compartmentalized DNA storage compared with repeated random access and reduces amplification bias tenfold during multiplex polymerase chain reaction. Using fluorescent sorting, we also demonstrate sample pooling and data retrieval by microcapsule barcoding. Therefore, the thermoresponsive microcapsule technology offers a scalable, sequence-agnostic approach for repeated random access to archival DNA files.“














 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

