From Tianjin University, Exogenous Artificial DNA Forms Chromatin Structure With Active Transcription in Yeast
Work is key proof-of-concept that demonstrated how artificial chromosomes can be used for storage in way that is robust and essentially free to copy.
This is a Press Release edited by StorageNewsletter.com on January 21, 2022 at 2:01 pmPreviously, researchers from Tianjin University, China, designed and created a 254 kb digital data carrying artificial chromosome in yeast.
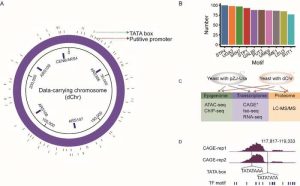
Genetic elements in data-carrying DNA.
(Credit: © Science China Press)
Click to enlarge
This work ‘is a key proof-of-concept that demonstrated how artificial chromosomes can be used for data storage in a way that is robust and essentially free to copy’. It broke through the limitation that the total length of DNA carrying encoded data within a single cell was just a few thousand base pairs per genome (X. Lu and T. Elli,National Science Review 2021 Vol. 8 Issue 7, DOI:10.1093/nsr/nwab086). The intracellular biological characteristics of this completely exogenous digital data-carrying chromosome (dChr) have received widespread attention, such as its genetic and epigenetic features in living cells.
The dChr was designed for data storage purpose. Researchers applied the superposition method with sparsified low-density parity-check (LDPC) codes and pseudo-random sequences to encode the image information. No extra caution was taken into consideration to avoid introducing potential genetic elements with biological activities such as promoters, regulatory sequences, protein/RNA binding and coding sequences.
“Many genetic elements were generated unintentionally. We found 54 putative promoters, 36 TATA boxes, and a large number of transcription factor-binding motifs that were distributed in the dChr, ” said Dr. Jianting Zhou, first author for this work. By using CAGE-seq (Cap Analysis of Gene Expression AND deep Sequencing), 20 de novo TSSes (Transcriptional Start Sites) in the dChr were detected, as well as TATA boxes near them.
The above data suggested that there are TSSes mixed into DNA sequence of the dChr. PolII may bind to these TSSes and further activate the transcription machine, producing stable mRNAs. The results also consistent with the high H3K4 tri-methylation modification and chromatin accessibility data in the dChr. The dChr formed chromatin with histones in vivo, and the artificial chromatin displayed an active epigenetic state with high chromatin accessibility and H3K4me3 levels. As for three-dimensional structure of the dChr, the researchers inspected the trans-interaction matrix and confirmed the yeast chromosomes exhibited a Rabl configuration.
These encouraging results confirmed that the exogenous dChr was not naked DNA in yeast, and it wrapped around histones, contributing to the formation of chromatin with three-dimensional structures. This study deepens our understanding of the activity of the fully exogenous YAC in the yeast cells, and guides researchers to construction of next generation of artificial chromosomes for data storage.
This work was supported by grants from the National Key R&D Program of China (2121YFA0909300), the National Natural Science Foundation of China (31861143017, 21621004, and 31901019), and the China Postdoctoral Science Foundation (2021M692389).
Article: Exogenous artificial DNA forms chromatin structure with active transcription in yeast
Science China Life Sciences Journal has published an article written by Jianting Zhou, Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, 300072, China, and SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300072, China, Chao Zhang, Ran Wei, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing, 100871, China, Mingzhe Han, Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, 300072, China, and SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300072, China, Songduo Wang, CAS Key Laboratory of Separation Science for Analytical Chemistry, National Chromatographic R. & A. Center, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, 116023, China, and University of Chinese Academy of Sciences, Beijing, 100049, China, Kaiguang Yang, Lihua Zhang, CAS Key Laboratory of Separation Science for Analytical Chemistry, National Chromatographic R. & A. Center, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, 116023, China , Weigang Chen, School of Microelectronics, Tianjin University, Tianjin, 300072, China, Mingzhang Wen, Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, 300072, China, and SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300072, China, Cheng Li, Wei Tao, The MOE Key Laboratory of Cell Proliferation and Differentiation, School of Life Sciences, Peking University, Beijing, 100871, China, Ying-Jin Yuan, Frontier Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin, 300072, China, and SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), School of Chemical Engineering and Technology, Tianjin University, Tianjin, 300072, China.
Abstract: “Yeast artificial chromosomes (YACs) are important tools for sequencing, gene cloning, and transferring large quantities of genetic information. However, the structure and activity of YAC chromatin, as well as the unintended impacts of introducing foreign DNA sequences on DNA-associated biochemical events, have not been widely explored. Here, we showed that abundant genetic elements like TATA box and transcription factor-binding motifs occurred unintentionally in a previously reported data-carrying chromosome (dChr). In addition, we used state-of-the-art sequencing technologies to comprehensively profile the genetic, epigenetic, transcriptional, and proteomic characteristics of the exogenous dChr. We found that the data-carrying DNA formed active chromatin with high chromatin accessibility and H3K4 tri-methylation levels. The dChr also displayed highly pervasive transcriptional ability and transcribed hundreds of noncoding RNAs. The results demonstrated that exogenous artificial chromosomes formed chromatin structures and did not remain as naked or loose plasmids. A better understanding of the YAC chromatin nature will improve our ability to design better data-storage chromosomes. “













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

