Northwestern University Says: “Our DNA is Becoming World’s Tiniest Hard Drive“
Researchers propose faster method for recording data to DNA, showing promise in fields of storage, neuron recording, work expands the repertoire of DNA-based recording techniques by developing a novel DNA synthesis-based system that can record temporal environmental signals into DNA with resolution of minutes
This is a Press Release edited by StorageNewsletter.com on October 13, 2021 at 1:32 pmFrom Northwestern University
Summary:
-
Current methods using DNA to store data inside or outside cell are costly and take days to produce recordings
-
New method works more than 10x faster than other intracellular solutions, transferring information to DNA in seconds instead of hours
-
Researchers believe their format of DNA data could be used to record neuron behavior, uncovering mysteries about what happens in the brain
Our genetic code is millions of times more efficient at storing data than existing solutions, which are costly and use amounts of energy and space. In fact, we could get rid of HDDs and store all the digital data on the planet within a couple hundred pounds of DNA.
Using DNA as a high-density data storage medium holds the potential to forge breakthroughs in biosensing and biorecording technology and next-gen digital storage, but researchers haven’t been able to overcome inefficiencies that would allow the technology to scale.
Now, researchers at Northwestern University propose a new method for recording information to DNA that takes minutes, rather than hours or days, to complete. The team used a novel enzymatic system to synthesize DNA that records rapidly changing environmental signals directly into DNA sequences, a method the paper’s senior author said could change the way scientists study and record neurons inside the brain.
The research, Recording Temporal Signals with Minutes Resolution Using Enzymatic DNA Synthesis, was published September 30, 2021) in the Journal of the American Chemical Society.
The paper’s senior author, Northwestern engineering professor Keith E.J. Tyo, said his lab was interested in leveraging DNA’s natural abilities to create a new solution for storing data.
“Nature is good at copying DNA, but we really wanted to be able to write DNA from scratch,” Tyo said. “The ex vivo (outside the body) way to do this involves a slow, chemical synthesis. Our method is much cheaper to write information because the enzyme that synthesizes the DNA can be directly manipulated. State-of-the-art intracellular recordings are even slower because they require the mechanical steps of protein expression in response to signals, as opposed to our enzymes which are all expressed ahead of time and can continuously store information.”
Tyo, professor, chemical and biological engineering, McCormick School of Engineering, is a member of Center for Synthetic Biology, and studies microbes and their mechanisms for sensing environmental changes and responding to them quickly.
Bypassing protein expression
Existing methods to record intracellular molecular and digital data to DNA rely on multipart processes that add new data to existing sequences of DNA. To produce an accurate recording, researchers must stimulate and repress expression of specific proteins, which can take over 10 hours to complete.
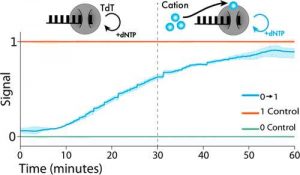
The Tyo lab hypothesized they could use a new method that they called Time-sensitive Untemplated Recording using Tdt for Local Environmental Signals, or TURTLES, to synthesize completely new DNA instead of copying a template of it, making a faster and higher resolution recording.
As the DNA polymerase continues to add bases, data is recorded into the genetic code on a scale of minutes as changes in the environment impact the composition of the DNA it synthesizes. The environmental changes, such as changes in the concentration of metals, are recorded by the polymerase, acting as a ‘molecular ticker tape’ and indicating to scientists the time of an environmental change. Using biosensors to record changes into DNA represents a major step in proving TURTLES’ viability for use inside cells, and could give researchers the ability to use recorded DNA to learn about how neurons communicate with each other.
“This is a really exciting proof of concept for methods that could one day lets us study the interactions between millions of cells simultaneously,” said Namita Bhan, co-first author and postdoctoral researcher, Tyo lab. “I don’t think there’s any previously reported direct enzyme modulation recording system.”
From brain cells to polluted water
With more potential for scalability and accuracy, TURTLES could offer the basis for tools that catapult brain research forward. According to Alec Callisto, also a co-first author and graduate student in the Tyo lab, researchers can only study a tiny fraction of a brain’s neurons with today’s technology, and even then, there are limits on what they know they do. By placing recorders inside all the cells in the brain, scientists could map responses to stimuli with single-cell resolution across many (million) neurons.
“If you look at how current technology scales over time, it could be decades before we can even record an entire cockroach brain simultaneously with existing technologies – let alone the tens of billions of neurons in human brains,” Callisto said. “So that’s something we’d really like to accelerate.”
Outside the body, the TURTLES system also could be used for a variety of solutions to address the explosive growth in data storage needs (up to 175ZB by 2025).
It’s particularly good for long term archival data applications such as storing closed-circuit security footage, which the team refers to as data that you ‘write once and read never,’ but need to have accessible in the event an incident occurs. With technology developed by engineers, hard drives and disk drives that hold years of beloved camera memories also could be replaced by bits of DNA.
Outside of storage, the ‘ticker tape’ function could be used as a biosensor to monitor environmental contaminants, like the heavy metal concentration in drinking water.
While the lab focuses on moving beyond a proof of concept in both digital and cellular recording, the team expressed hope that more engineers would take interest in the concept and be able to use it to record signals important to their research.
“We’re still building out the genomic infrastructure and cellular techniques we need for robust intracellular recording,” Tyo said. “This is a step along the way to getting to our long-term goal.”
This work was funded by two National Institutes of Health grants (R01MH103910; and UF1NS107697) and an NIH Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. The research was supported in part through the computational resources and staff contributions provided for the Quest high-performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research and Northwestern University Information Technology. All next-generation sequencing was done with the help of the Next Generation Sequencing Core facility at the University of Illinois at Chicago. Sanger sequencing was supported by the Northwestern University NUSeq Core Facility. Gel imaging was supported by the Northwestern University Keck Biophysics Facility and a Cancer Center Support Grant (NCI CA060553). The Keck Biophysics Facility’s Azure Sapphire Imager was funded by an NIH grant (1S10OD026963-01). Protein purification was supported by the Northwestern University Recombinant Protein Production Core.
Article: Recording Temporal Signals with Minutes Resolution Using Enzymatic DNA Synthesis
Journal of the American Chemical Society has published an article written by Namita Bhan, Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, United States, and Mitolab, Cambridge, Massachusetts 02139, United States, Alec Callisto, Jonathan Strutz, Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, United States, Joshua Glaser, Center for Theoretical Neuroscience, Columbia University, New York, New York 10027, United States, Reza Kalhor, Department of Biomedical Engineering, Center for Epigenetics, Johns Hopkins School of Medicine, Baltimore, Maryland 21205, United States, Edward S. Boyden, Department of Brain and Cognitive Sciences, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States, McGovern Institute, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, United States, and Howard Hughes Medical Institute, Department of Neurobiology, Harvard Medical School, Boston, Massachusetts 02115, United States, George Church, Department of Biomedical Engineering, Center for Epigenetics, Johns Hopkins School of Medicine, Baltimore, Maryland 21205, United States, Konrad Kording, Department of Neuroscience, University of Pennsylvania, Philadelphia, Pennsylvania 19104, United States, and Keith E. J. Tyo, Department of Chemical and Biological Engineering, Northwestern University, Evanston, Illinois 60208, United States.
Abstract: “Employing DNA as a high-density data storage medium has paved the way for next-generation digital storage and biosensing technologies. However, the multipart architecture of current DNA-based recording techniques renders them inherently slow and incapable of recording fluctuating signals with subhour frequencies. To address this limitation, we developed a simplified system employing a single enzyme, terminal deoxynucleotidyl transferase (TdT), to transduce environmental signals into DNA. TdT adds nucleotides to the 3′-ends of single-stranded DNA (ssDNA) in a template-independent manner, selecting bases according to inherent preferences and environmental conditions. By characterizing TdT nucleotide selectivity under different conditions, we show that TdT can encode various physiologically relevant signals such as Co2+, Ca2+, and Zn2+ concentrations and temperature changes in vitro. Further, by considering the average rate of nucleotide incorporation, we show that the resulting ssDNA functions as a molecular ticker tape. With this method we accurately encode a temporal record of fluctuations in Co2+ concentration to within 1 min over a 60 min period. Finally, we engineer TdT to allosterically turn off in the presence of a physiologically relevant concentration of calcium. We use this engineered TdT in concert with a reference TdT to develop a two-polymerase system capable of recording a single-step change in the Ca2+ signal to within 1 min over a 60 min period. This work expands the repertoire of DNA-based recording techniques by developing a novel DNA synthesis-based system that can record temporal environmental signals into DNA with a resolution of minutes.“













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter
