For German Climate Computing Center, StrongBox Building Exabyte Data Archive to Advance Climate Science Research
Leveraging power of metadata for global access to all data across all storage types
This is a Press Release edited by StorageNewsletter.com on April 6, 2021 at 2:30 pmBy Carsten Schmitt, storage administrator, Deutsches Klimarechenzentrum GmbH (DKRZ)
The German Climate Computing Center (DKRZ) is a central service center for German climate and earth system research and provides infrastructure for simulation-based climate science.
Researchers from all over the world access this shared system for compute and data services to advance climate science research. A significant portion of these services include the management of storage and archives, with over 150PB of active archive data, which is growing rapidly and with the new HPC cluster, that will be installed this year, the data archive storage is expected to support a growth rate of at least 120PB per year.
A common problem in climate science is that data is often scattered over multiple storage resources, and climate scientists need to spend a lot of time gathering data from different data sources before they can even begin their work.
DKRZ’s new ExabyteB Data Archive project addresses this problem by providing researchers with new metadata-driven tools based upon StrongLink data management software from StrongBox Data Solutions, Inc. to find and manage their data more easily across both existing and new multi-tiered storage platforms. This new system also provides the foundation for efficiently managing the anticipated data growth, and also the expanding multi-protocol access requirements needed for new workflow models in the future.
As part of this project, DKRZ is also modernizing its tiered architecture to move from proprietary HSM tape formats to open standard LTFS format, so data can remain open and not bound to one company, and in this way will remain available for scientists for years to come. This large-scale transition from proprietary data formats to open standards is metadata-driven, meaning that the migration process of the 150PB of data on tape is designed to be seamless, and transparent to users, only requiring a very short downtime not typical for large-scale migrations of this type.
Enabling researchers global access to data
Scientists and scientific projects need to access and store multiple Petabytes of data. Typical workflows don’t require the data all at once, but any part of a project data should be immediately accessible to scientists with the shortest delay possible. This is not only a technical problem but also a problem of storage organization and data management. As data volumes grow at increasing rates, the problem gets more challenging as more and different storage types are needed at different phases of the data’s life cycle. This includes high performance HPC storage, to other tiers of nearline and tape-based active archive storage, across different gens of storage platforms in each category.
Some projects require data to be moved from the HPC system to the archive for some weeks to free up the high-performance storage, which has more restrictive quotas. When the data needs to be brought back to the HPC system to process new calculations, then it is copied back to the cluster. In this regard the tape archive is more used as an external cache and users should be able to move these data conveniently between the different storage systems.
But the main purpose of the Exabyte Data Archive is to provide active storage capacity for research projects that often consume multiple petabytes for the duration of the project. And such projects can often last for multiple years, during which time the scientists need reliable and immediate access to the data for further calculations or publications. Depending on the reproducibility, some data like sensor data or very expensive calculations may have to be stored with extra copies at an external site.
In addition to such long-term active access to the project data, even when some of the projects are finalized the data will be archived for ten years and if needed must be retrievable at any time. Data integrity is extremely important, and it must stay unchanged. Although a researcher’s initial project may be complete, dataset copies from such projects may be used for additional projects, or the research results may need to be available for review by others at a later time. To fulfill this requirement, the data is stored with additional copies and the integrity of the data and the storage media must be assured as the highest priority in the design requirements for the Exabyte Data Archive.
The challenge has been how to provide researchers with global access to their data across any of the storage tiers and multiple storage types from different vendors, so they can focus their efforts on research activities, and not on wasting time trying to locate datasets across different storage silos. This additional workload for researchers can partly be avoided by providing global access to commonly used climate data or results of standard calculations stored in the tape archive, so that they can also access the prepared data from the HPC cluster.
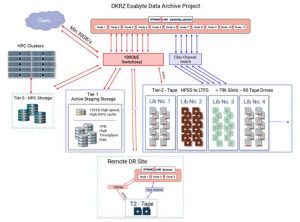
Fig. 1: High-level architecture of the DKRZ Exabyte Data Archive system,
which includes 1PB of archive disk cache, plus 79,000 slots across
multiple vendor tape libraries. The StrongLink metadata database and a selected
subset of critical data are automatically replicated to a second site continually for DR protection.
Click to enlarge
Main goals of new Exabyte Data Archive
The many different use cases needed by scientists require certain functions for data movement, storage and retrieval. At DKRZ we are planning to provide these functions for users and administrators alike with the help of the StrongLink software.
On many HPC clusters some form of Linux is used as the OS especially in a scientific environment. Therefore most of the time data transfers from and to the archive are part of an automated process either via a shell or with a cluster resource manager on compute nodes. DKRZ needed a command line tool that could be integrated in interactive and non-interactive workflows, so that researchers could have easy access to the global namespace storage environment that spans multiple storage platforms and vendors. StrongLink includes a command line utility to provide this capability, in addition to other metAdata: query and other functions noted below.
Because a lot of the data transfers are happening automatically by policy, every transfer to and within the data archive can be checked with automatically generated checksums by StrongLink. The integrity of the data during and after transfers is one of the main concerns of the scientists, and they don’t want to spend their time managing these integrity checks themselves.
Another factor is that additional to the established data flows to the archive there can be the need to expand the workflows to include other storage types. For example, in cases where large project datasets from another external source need to be integrated with a one-time import into the data archive, to be used locally with existing datasets. The ability to easily connect additional often incompatible storage sources and targets to the Stronglink Cluster without the need to change the main setup and possibly disturbing the running workflows is another positive aspect, and it will enable us to use available storage resources more efficiently.
Transitioning to LTFS without disruption for researchers
As described above, the research data on tape must be available for many years. This need for longevity requires a storage format that is open and not bound to one company. This was the rationale for transitioning the tape archive from a proprietary data format to open-standard LTFS. This should ensure accessibility and stability of the tape data for the coming years, independent from a specific vendor. In addition, LTFS format ensures that data tapes may be exported and shared with other research organizations without the dependency on StrongLink, or any other system.
The challenge for large-scale data migrations of this type between incompatible systems or formats is to make sure that users are not impacted by any changes to their workflow during the transition. Researchers need to have a single predictable workflow that can enable them to see all of the data in a global namespace, without the added burden of needing to figure out whether it is on the legacy system or in the new format. In the current DKRZ environment, there are 4 tape libraries from different vendors at the primary datacenter, with 79,000 slots and 90 tape drives. But even with that much throughput, migrating 150PB from a proprietary format to LTFS will take time. In addition to the migration, we anticipate that the same libraries will need to support at least 120PB of I/O per year for day-to-day researcher workflows.
To ensure that researchers are not hindered by the transition, StrongLink will aggregate metadata from the legacy system so that scientists will be provided with a single global view to all 150PB of data, regardless of whether the files are on legacy tape format, or on new LTFS format. In the background StrongLink will be migrating data from the legacy tapes to repack and write back in LTFS, but users will not be aware of this.
From a user’s perspective, the workflow will be as it is today, which is typically via CLI access to the archive. The difference is that this will now be done with the StrongLink CLI utility, and will give them global access to all the data in both legacy and LTFS formats. When a scientist initiates a data recall, StrongLink will read the data from either format, and move the data to HPC storage or other target storage. When the researcher has finished the HPC run and moves the resulting datasets back to the archive, the system will automatically write to tape in the new LTFS format. The scientist does not need to know about this back-end orchestration.
StrongLink’s migration policy will prioritize user-initiated I/O over the background tape migration jobs. During downtime, StrongLink will take over all 90 tape drives to accelerate the migration. But when users initiate a data recall from tape, the system will automatically pause the migration jobs to enable user workflows to take over the drives it needs.
Using this methodology, the 150PB migration will be effectively accomplished with less than one-week downtime for the user, even as the physical migration takes place over time in the background.
Enabling richer metadata management to extend utilization of archive
StrongLink allows users to create custom metadata for any file or dataset, which greatly expands the capabilities for the data management. These may be simple metadata tags, or multi-value metadata forms, which should help scientists to solve many problems with data organization. Until now, often such metadata is stored as part of the file name and the path in the POSIX file system, and this can be problematic if paths or file names change either by accident or by organizational restructuring. In addition, researchers may keep such metadata in external databases that are disconnected from the actual files. When the scientists are enabled to add metadata to the files for easier searches and data retrieval, a lot of time and work can be saved.
Another advantage of such individual metadata is to use them in automated workflows. Some data movement, like the long term archive, is still only partly automated. But with StrongLink metadata aggregation, workflow automation may be triggered by any combination of metadata variables, including custom user-created metAdata: POSIX metadata from the file systems, or rich file metadata such as NetCDF. With these additional metadata capabilities, we want to be able to create fully automated workflows. This should also help us to create reproducible and standardized data movements to minimize the chance for errors.
Preparing for future
The Exabyte Data Archive project is preparing the way for the future of climate science research at DKRZ by scientists from all over the world. It is a system designed to provide researchers with maximum efficiency for data access and data management across what is inevitably a large and heterogeneous storage environment. The system prioritizes the needs of the researchers to focus on their scientific work, and seeks to minimize the time needed to find or manage data through greater automation and global access across all current and future storage resources. This is key to providing the foundation for the significant growth in research activities and data in the future.














 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter
