R&D: Bacterial Nanopores Open Future of Storage From EPFL/University of Strasbourg
Findings open promising possibilities to develop writing-reading technologies to process digital data using a biological-inspired platform.
This is a Press Release edited by StorageNewsletter.com on December 17, 2020 at 2:10 pmFrom EPFL (École Polytechnique Fédérale de Lausanne)
Bioengineers at EPFL (École Polytechnique Fédérale de Lausanne) have developed a nanopore-based system that can read data encoded into synthetic macromolecules with higher accuracy and resolution than similar methods on the market.
Engineered bacterial pores can decode digital information stored
in tailored-made polymers.
(Credit: Matteo Dal Peraro (aerolysin structure)/iStock background)

The system is also potentially cheaper and longer-lasting, and overcomes limitations that prevent us from moving away from conventional data storage devices that are rapidly maxing out in capacity and endurance.
In 2020, each person in the world is producing about 1.7MB of data every second. In just a single year, that amounts to 418ZB – or 418 billion one-terabyte hard drives.
We currently store data as 1s and 0s in magnetic or optical systems that don’t last a century. Meanwhile, data centers consume massive amounts of energy and produce enormous carbon footprints. Simply put, the way we store our ever-growing volume of data is unsustainable.
DNA as data storage
But there is an alternative: storing data in biological molecules such as DNA. In nature, DNA encodes, stores, and makes readable massive amounts of genetic information in tiny spaces (cells, bacteria, viruses) – and does so with a high degree of safety and reproducibility.
Compared to conventional data-storage devices, DNA is more enduring and compacted, can retain ten times more data, has a million-fold higher storage density, and consumes 100 million times less energy to store the same amount of data as a drive. Also, a DNA-based data-storage device would be tiny: a year’s worth of global data can be stored in just four grams of DNA.
But storing data with DNA also involves exorbitant costs, painfully slow writing and reading mechanisms, and is susceptible to mis-readings.
Nanopores to the rescue
A way is to use nano-sized holes called nanopores, which bacteria often punch into other cells to destroy them. The attacking bacteria use specialized proteins known as ‘pore-forming toxins’ which latch onto the cell’s membrane and form a tube-like channel through it.
In bioengineering, nanopores are used for ‘sensing’ biomolecules, such as DNA or RNA. The molecule passes through the nanopore like a string, steered by voltage, and its different components produce distinct electrical signals (an ‘ionic signature’) that can be used to identify them. And because of their high accuracy, nanopores have also been tried out for reading DNA-encoded information.
Nonetheless, nanopores are still limited by low-resolution readouts – a real problem if nanopore systems are ever to be used for storing and reading data.
Aerolysin nanopores
The potential of nanopores inspired scientists at EPFL’s School of Life Sciences to explore nanopores produced by the pore-forming toxin aerolysin, made by the bacterium Aeromonashydrophila. Led by Matteo Dal Peraro, School of Life Sciences, EPFL, the researchers show that aerolysin nanopores can be used for decoding binary information.
In 2019, Dal Peraro’s lab showed that nanopores can be used for sensing more complex molecules, like proteins. In this study, published in Science Advances, the team joined force with the lab of Alexandra Radenovic (EPFL School of Engineering) and adapted aerolysin to detect molecules tailored-made precisely to be read by this pore. The technology has been filed as a patent.
The molecules, known as ‘digital polymers’, were developed in the lab of Jean-François Lutz, Institut Charles Sadron, CNRS, in Strasbourg. They are a combination of DNA nucleotides and non-biological monomers designed to pass through aerolysin nanopores and give out an electrical signal that could be read out as a ‘bit’.
The researchers used aerolysin mutants to systematically design nanopores for reading out signals of their informational polymers. They optimized the speed of the polymers passing through the nanopore so that it can give out a uniquely identifiable signal. “But unlike conventional nanopore readouts, this signal delivered digital reading with single-bit resolution, and without compromising information density,” says Dr Chan Cao, first author of the paper.
To decode the readout signals the team used deep learning, which allowed them to decode up to 4 bits of information from the polymers with high accuracy. They also used the approach to blindly identify mixtures of polymers and determine their relative concentration.
The system is considerably cheaper than using DNA for data-storage, and offers longer endurance. In addition, it is ‘miniaturizable’, meaning that it could easily be incorporated into portable data-storage devices.
“There are several improvements we are working on to transform this bio-inspired platform into an actual product for data storage and retrieval,” says Matteo Dal Peraro. “But this work clearly shows that a biological nanopore can read hybrid DNA-polymer analytes. We are excited as this opens up new promising perspectives for polymer-based memories, with important advantages for ultrahigh density, long-term storage and device portability.”
Other contributor:
University of Strasbourg
Article: Aerolysin nanopores decode digital information stored in tailored macromolecular analytes
Science Advances has published an article written by Chan Cao, Lucien F. Krapp, Institute of Bioengineering, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland, and Swiss Institute of Bioinformatics (SIB), 1015 Lausanne, Switzerland, Abdelaziz Al Ouahabi, Niklas F. König, Université de Strasbourg, Centre national de la recherche scientifique (CNRS), Institute Charles Sadron UPR22, 23 rue du Loess, 67034 Strasbourg Cedex 2, France, Nuria Cirauqui, Institute of Bioengineering, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland, Department of Pharmaceutical Biotechnology, Universidade Federal do Rio de Janeiro, 21941-902 Rio de Janeiro, Brazil, and CNRS, UMR5086, “Molecular Microbiology and Structural Biochemistry”, University of Lyon, 7 Passage du Vercors, 69367 Lyon, France, Aleksandra Radenovic, Institute of Bioengineering, School of Engineering, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland, ean-François Lutz, Université de Strasbourg, Centre national de la recherche scientifique (CNRS), Institute Charles Sadron UPR22, 23 rue du Loess, 67034 Strasbourg Cedex 2, France, and Matteo Dal Peraro, Institute of Bioengineering, School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015 Lausanne, Switzerland, and Swiss Institute of Bioinformatics (SIB), 1015 Lausanne, Switzerland.
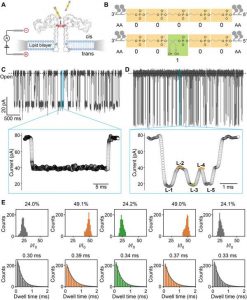
Aerolysin reading of polymers encoding single-bit information.
(A) Illustration of single-channel recording setup using an aerolysin pore; the cis and trans chambers are filled with 1.0 M KCl electrolyte buffer, and voltage is applied across the pore using two Ag/AgCl electrodes. Amino acid at 220 and 238 position are highlighted by red and blue, respectively. (B) Schematic structure of two representative polymers: AA00000AA and AA00100AA. (C) Raw current trace of AA00000AA during single-channel recording measurement (top). Magnification of one single event (bottom). (D) Raw current trace of AA00100AA measurement. Magnification of one single event (bottom) showing a multilevel signal: L-1 (gray), L-2 (orange), L-3 (green), L-4 (orange), and L-5 (gray). (E) I/I0 histogram (top) and dwell time distribution (bottom) for L-1, L-2, L-3, L-4, and L-5, respectively. Relative fitted values are reported in each figure. All data were obtained using 1.0 M KCl, 10 mM tris, and 1.0 mM EDTA at pH 7.4, applying a bias potential of 100 mV.
Abstract: “Digital data storage is a growing need for our society and finding alternative solutions than those based on silicon or magnetic tapes is a challenge in the era of ‘big data.’ The recent development of polymers that can store information at the molecular level has opened up new opportunities for ultrahigh density data storage, long-term archival, anticounterfeiting systems, and molecular cryptography. However, synthetic informational polymers are so far only deciphered by tandem mass spectrometry. In comparison, nanopore technology can be faster, cheaper, nondestructive and provide detection at the single-molecule level; moreover, it can be massively parallelized and miniaturized in portable devices. Here, we demonstrate the ability of engineered aerolysin nanopores to accurately read, with single-bit resolution, the digital information encoded in tailored informational polymers alone and in mixed samples, without compromising information density. These findings open promising possibilities to develop writing-reading technologies to process digital data using a biological-inspired platform.“














 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

