R&D: DNA Punch Cards for Storing Data on Native DNA Sequences Via Enzymatic Nicking
Use Pyrococcus furiosus Argonaute to punch files into PCR products of Escherichia coli genomic DNA and accurately reconstruct encoded data through high-throughput sequencing and read alignment.
This is a Press Release edited by StorageNewsletter.com on April 24, 2020 at 2:23 pmNature Communications has published an article written by S. Kasra Tabatabaei, Center for Biophysics and Quantitative Biology, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Boya Wang, Department of Electrical and Computer Engineering, University of Texas at Austin, Austin, TX, 78712, USA, Nagendra Bala Murali Athreya, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Behnam Enghiad, Department of Chemical and Biomolecular engineering, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Alvaro Gonzalo Hernandez, Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Christopher J. Fields, High Performance Computing in Biology (HPCBio), Roy J. Carver Biotechnology Center, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Jean-Pierre Leburton, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, David Soloveichik, Department of Electrical and Computer Engineering, University of Texas at Austin, Austin, TX, 78712, USA, Huimin Zhao, Center for Biophysics and Quantitative Biology, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, Department of Chemical and Biomolecular engineering, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, and Carl R. Woese Institute for Genomic Biology, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA, and Olgica Milenkovic, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, 61801, USA.
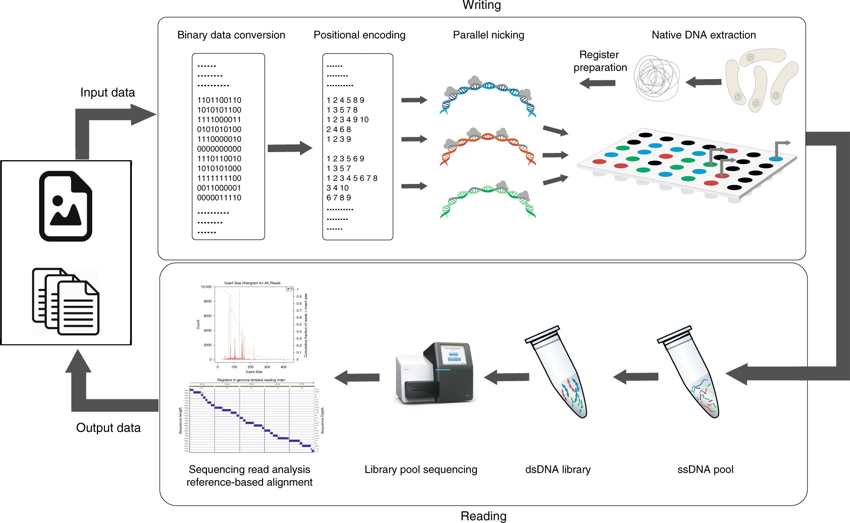
The native DNA-based data storage platform.
In the Write component, arbitrary user content is converted into a binary message. The message is then parsed into blocks of m bits, where m corresponds to the number of nicking positions on the register (for the running example, m = 10). Different (orthogonal) registers may be used to encode information in parallel, as indicated by the different colors of the DNA strings. Subsequently, binary information is translated into positional information indicating where to nick. Nicking reactions are performed in parallel via combinations of PfAgo and guides. In the Read component, nicked products are purified and denatured to obtain a pool of ssDNAs of different lengths. The pool of ssDNAs is sequenced via MiSeq either as part of an orthogonal register mixture or individually. The output reads are processed by first performing reference-based alignment of the reads, and then using read coverages to determine the nicked positions.
Abstract: “Synthetic DNA-based data storage systems have received significant attention due to the promise of ultrahigh storage density and long-term stability. However, all known platforms suffer from high cost, read-write latency and error-rates that render them noncompetitive with modern storage devices. One means to avoid the above problems is using readily available native DNA. As the sequence content of native DNA is fixed, one can modify the topology instead to encode information. Here, we introduce DNA punch cards, a macromolecular storage mechanism in which data is written in the form of nicks at predetermined positions on the backbone of native double-stranded DNA. The platform accommodates parallel nicking on orthogonal DNA fragments and enzymatic toehold creation that enables single-bit random-access and in-memory computations. We use Pyrococcus furiosus Argonaute to punch files into the PCR products of Escherichia coli genomic DNA and accurately reconstruct the encoded data through high-throughput sequencing and read alignment.“













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

