R&D: Characterization of DNA Storage Channel
Study to help guide design of future DNA storage systems by providing quantitative and qualitative understanding of DNA storage channel
This is a Press Release edited by StorageNewsletter.com on July 16, 2019 at 1:53 pmNature Scientific Reports has published an article written by Reinhard Heckel, Rice University, Department of Electrical and Computer Engineering, Houston, 77005, Texas, USA, Gediminas Mikutis, and Robert N. Grass, ETH Zurich, Department of Chemistry and Applied Biosciences, Zurich, 8093, Switzerland.
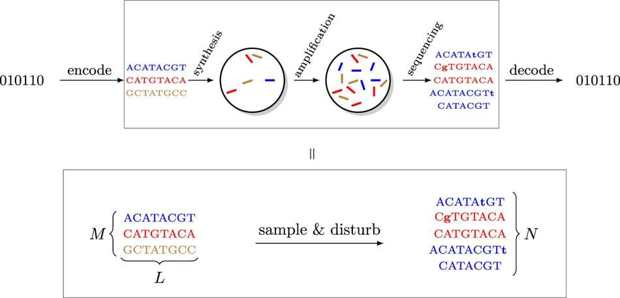
Channel model for DNA storage systems. Only short molecules can be synthesized, and of each molecule a large number of copies is generated in the synthesis process. For reading, the data is first amplified and then sequenced. Abstractly, the input to the channel is a multi-set of M length-L DNA molecules, while the output is a multi-set of N draws from the pool of DNA molecules that is disturbed by insertions, substitutions, and deletions (marked as lowercase and boldface letters). The sampling distribution as well as the process inducing errors in individual molecules account for errors in synthesis, storage, and sequencing.
Abstract: “Owing to its longevity and enormous information density, DNA, the molecule encoding biological information, has emerged as a promising archival storage medium. However, due to technological constraints, data can only be written onto many short DNA molecules that are stored in an unordered way, and can only be read by sampling from this DNA pool. Moreover, imperfections in writing (synthesis), reading (sequencing), storage, and handling of the DNA, in particular amplification via PCR, lead to a loss of DNA molecules and induce errors within the molecules. In order to design DNA storage systems, a qualitative and quantitative understanding of the errors and the loss of molecules is crucial. In this paper, we characterize those error probabilities by analyzing data from our own experiments as well as from experiments of two different groups. We find that errors within molecules are mainly due to synthesis and sequencing, while imperfections in handling and storage lead to a significant loss of sequences. The aim of our study is to help guide the design of future DNA data storage systems by providing a quantitative and qualitative understanding of the DNA data storage channel.“













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter


