R&D: Random Access in Large-Scale DNA Data Storage
Demonstrate viable, large-scale system for DNA data storage and retrieval
This is a Press Release edited by StorageNewsletter.com on March 5, 2018 at 2:28 pmNature Biotechnology has published an article written by Lee Organick, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, Siena Dumas Ang, Yuan-Jyue Chen, Microsoft Research, Redmond, Washington, USA, Randolph Lopez, Department of Bioengineering Department, University of Washington, Seattle, Washington, USA, Sergey Yekhanin, Konstantin Makarychev, Miklos Z Racz, Govinda Kamath, Parikshit Gopalan, Bichlien Nguyen, Microsoft Research, Redmond, Washington, USA, Christopher N Takahashi, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, Sharon Newman, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, Hsing-Yeh Parker, Cyrus Rashtchian, Microsoft Research, Redmond, Washington, USA, Kendall Stewart, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, Gagan Gupta, Robert Carlson, John Mulligan, Douglas Carmean, Microsoft Research, Redmond, Washington, USA, Georg Seelig, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, and Department of Electrical Engineering, University of Washington, Seattle, Washington, USA, Luis Ceze, Paul G. Allen School of Computer Science and Engineering, University of Washington, Seattle, Washington, USA, and Karin Strauss, Microsoft Research, Redmond, Washington, USA.
Click to enlarge
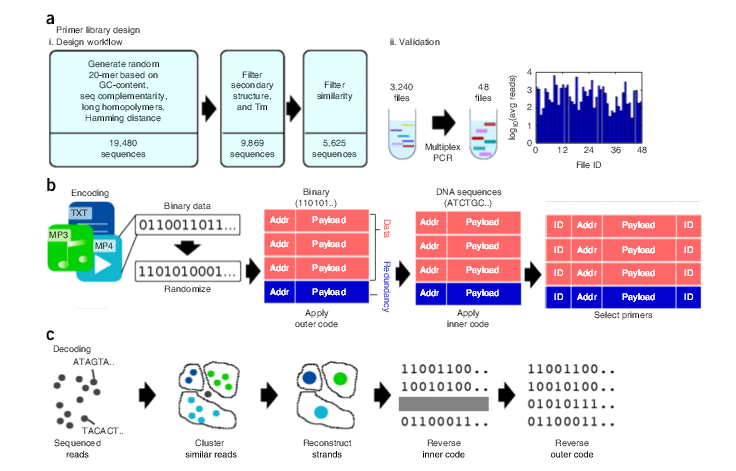
Abstract: “Synthetic DNA is durable and can encode digital data with high density, making it an attractive medium for data storage. However, recovering stored data on a large-scale currently requires all the DNA in a pool to be sequenced, even if only a subset of the information needs to be extracted. Here, we encode and store 35 distinct files (over 200 MB of data), in more than 13 million DNA oligonucleotides, and show that we can recover each file individually and with no errors, using a random access approach. We design and validate a large library of primers that enable individual recovery of all files stored within the DNA. We also develop an algorithm that greatly reduces the sequencing read coverage required for error-free decoding by maximizing information from all sequence reads. These advances demonstrate a viable, large-scale system for DNA data storage and retrieval.“













 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter

