R&D: DNA Fountain Enables Robust and Efficient Storage Architecture
Obtained retrieval from density of 215PB per gram of DNA
This is a Press Release edited by StorageNewsletter.com on March 10, 2017 at 2:31 pmScience Magazine has published an article written byYaniv Erlich, New York Genome Center, New York, NY 10013, USA, Department of Computer Science, Fu Foundation School of Engineering, Columbia University, New York, NY 10027, USA, and Center for Computational Biology and Bioinformatics (C2B2), Department of Systems Biology, Columbia University, New York, NY 10027, USA, and Dina Zielinski, New York Genome Center, New York, NY 10013, USA.
Reliable and efficient DNA storage architecture
DNA has the potential to provide large-capacity information storage. However, current methods have only been able to use a fraction of the theoretical maximum. Erlich and Zielinski present a method, DNA Fountain, which approaches the theoretical maximum for information stored per nucleotide. They demonstrated efficient encoding of information -including a full computer operating system – into DNA that could be retrieved at scale after multiple rounds of polymerase chain reaction.
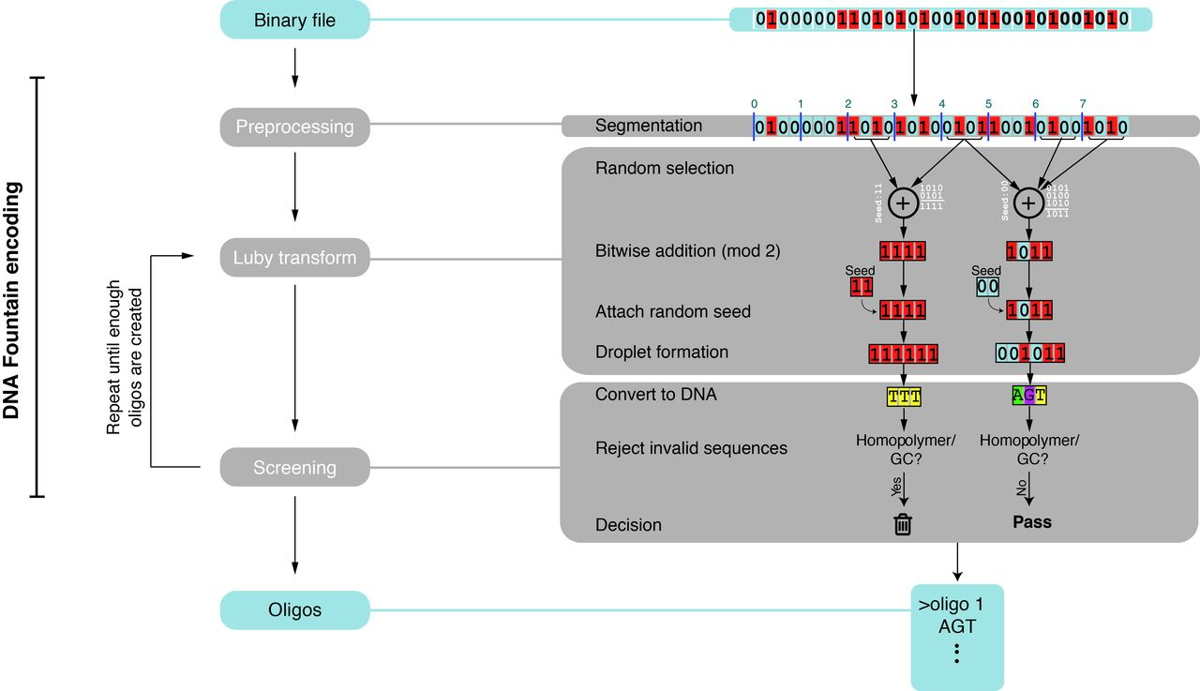
Fig. 1 DNA Fountain encoding.
(Left) Three main algorithmic steps. (Right) Example with a small file
of 32 bits. For simplicity, we partitioned the file into eight segments of 4 bits each.
The seeds are represented as 2-bit numbers and are presented for display
purposes only. See (12) for the full details of each algorithmic step.
Click to enlarge
Abstract: “DNA is an attractive medium to store digital information. Here we report a storage strategy, called DNA Fountain, that is highly robust and approaches the information capacity per nucleotide. Using our approach, we stored a full computer operating system, movie, and other files with a total of 2.14 × 106 bytes in DNA oligonucleotides and perfectly retrieved the information from a sequencing coverage equivalent to a single tile of Illumina sequencing. We also tested a process that can allow 2.18 × 1015 retrievals using the original DNA sample and were able to perfectly decode the data. Finally, we explored the limit of our architecture in terms of bytes per molecule and obtained a perfect retrieval from a density of 215PB per gram of DNA, orders of magnitude higher than previous reports.”












 Subscribe to our free daily newsletter
Subscribe to our free daily newsletter
